Exploring OkCupid Dataset with Machine Learning: Predicting Mating Success for Straight Males
🇨🇦From Data Preparation to Feature Engineering for Accurate Predictions
Book a callFrom Data Preparation to Feature Engineering for Accurate Predictions
Book a callFrom the previous blog article titled OkCupid Dataset Analysis for Machine Learning, we have decided to edit the choice of our features.
This project aims to create a predictive model by focusing exclusively on male entries from the dataset .
The target label will combine the "offspring" and "status" columns, which together capture mating success for males.
For the model's primary features, we will focus on "body_type," "ethnicity," and "height" as they may provide significant insights.
Additionally, secondary features such as "drinks," "drugs," "smokes," and "religion" will be incorporated to capture lifestyle and personal habits.
Finally, "income" may also be included as a tertiary feature to explore its potential influence, though it will have a lower priority in the analysis.
The aim is to carefully prepare and optimize this feature set to train a robust predictive model.
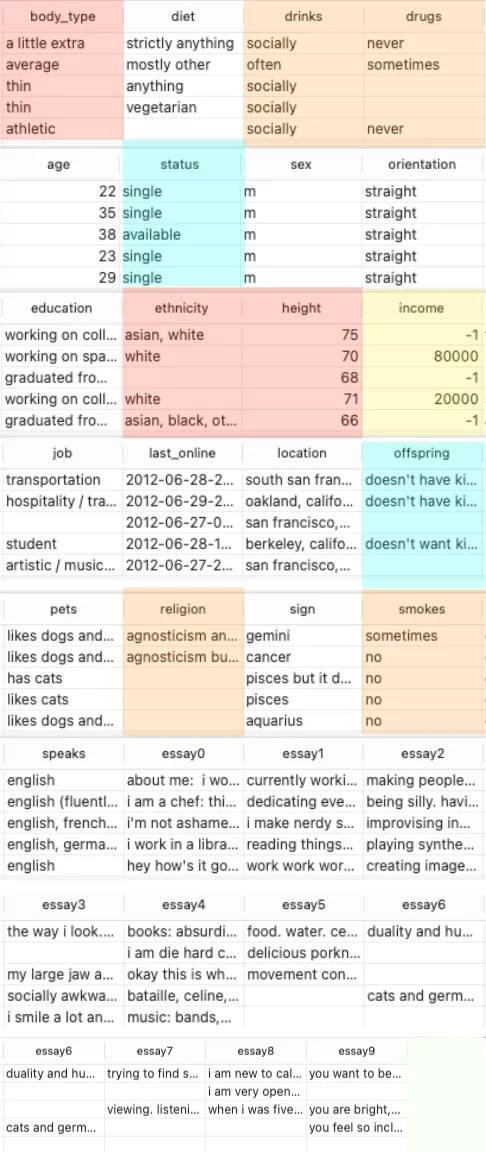
The OkCupid dataset contains various entries with different genders and orientations. Since our project aims to focus exclusively on mating success for straight males, the first step is to explore the available entries for the sex and orientation columns and filter out only those labelled as male and straight.
We will then store the filtered data in a new DataFrame and save it as a CSV file for further analysis.
The following Python code demonstrates how to inspect the available values in the sex and orientation columns and filter the data to keep only entries where the sex is "male" and the orientation is "straight."
./chapter2/data2/data_manipulations2/step1.py
dating_data["sex"].value_counts()
sex
m 35829
f 24117
Name: count, dtype: int64dating_data["orientation"].value_counts()
orientation
straight 51606
gay 5573
bisexual 2767
Name: count, dtype: int64import os
import pandas as pd
# Define the path to the dating dataset folder
DATING_PATH = os.path.join("datasets", "dating")
# Function to load the dating CSV file
def load_dating_data(dating_path=DATING_PATH):
csv_path = os.path.join(dating_path, "okcupid_profiles.csv") # Adjust the filename if necessary
return pd.read_csv(csv_path)
# Load the dating data
dating_data = load_dating_data()
# Filter the dataset to keep only male entries
male_data = dating_data[dating_data['sex'] == 'm']
# Further filter the dataset to keep only entries with orientation 'straight.'
male_straight_data = male_data[male_data['orientation'] == 'straight']
# Check the shape of the new filtered DataFrame to confirm the number of entries
print(f"Number of male and straight entries: {male_straight_data.shape[0]}")
# Save the filtered DataFrame to a new CSV file
output_path = "/Users/Zouhir/Documents/UQO_AUT2024/INF6333_code/book_aurelien_geron/chapter2/datasets/dating/step1_male_straight_data.csv"
male_straight_data.to_csv(output_path, index=False)
print(f"Filtered male and straight entries saved to: {output_path}")
Number of male and straight entries: 31073
Filtered male and straight entries saved to: /Users/Zouhir/Documents/UQO_AUT2024/INF6333_code/book_aurelien_geron/chapter2/datasets/dating/step1_male_straight_data.csvWhen working with machine learning, we typically divide the dataset into train and test sets:
However, datasets evolve. New entries might be added (e.g., more data collected), or some rows might be removed (e.g., duplicates or incorrect entries). If you don't have a stable way to split the data, your train and test sets could change between runs, leading to inconsistent results.
If your split is based on persistent IDs, deleting entries from the training set will not change the composition of the test set. The remaining test entries will stay the same as they were, ensuring no data leakage or unintended reassignment.
If new entries are added to the dataset (with their persistent IDs), they will be assigned to either the train or test set based on the same split logic. This ensures that the test set remains consistent, with only new data getting appropriately assigned.
One of the key benefits of using persistent IDs is that it guarantees consistent splits across dataset modifications.
Let's generate persistent IDs for our dataset:
./chapter2/data2/data_manipulations2/step2.py
import os
import pandas as pd
import uuid # To generate unique persistent IDs
# Define the path to the dataset folder
DATING_PATH = os.path.join("datasets", "dating")
# Function to load the dataset (male-only, straight)
def load_dating_data(dating_path=DATING_PATH):
csv_path = os.path.join(dating_path, "step1_male_straight_data.csv") # Ensure correct file extension
return pd.read_csv(csv_path)
# Load the male-straight-only dataset
male_data = load_dating_data()
# Create a persistent ID for each row if it doesn't already exist
if 'persistent_id' not in male_data.columns:
male_data['persistent_id'] = [str(uuid.uuid4()) for _ in range(len(male_data))]
print("Persistent IDs generated and assigned to all entries.")
# Save the updated dataset with the new persistent IDs
output_path = os.path.join(DATING_PATH, "male_straight_data_with_persistent_id.csv")
male_data.to_csv(output_path, index=False)
print(f"Dataset with persistent IDs saved to: {output_path}")
Persistent IDs are generated and assigned to all entries.
Dataset with persistent IDs saved to: datasets/dating/male_straight_data_with_persistent_id.csvNow that each entry has its respective and unique persistent ID, we can go ahead and execute the code below to split the data based on a 20% and 80% ratio for testing and training data, respectfully:
./chapter2/data2/train_test_data/script3.py
from zlib import crc32 # Import CRC32 hash function for consistency
import numpy as np
import pandas as pd
import os
# Define the path to the dataset folder
DATING_PATH = os.path.join("datasets", "dating")
# Function to load the dataset (male-only, straight, with persistent IDs)
def load_dating_data(dating_path=DATING_PATH):
csv_path = os.path.join(dating_path, "male_straight_data_with_persistent_id.csv")
return pd.read_csv(csv_path)
# Load the male-only data
male_straight_data_with_persistent_id = load_dating_data()
# Function to check if an identifier belongs in the test set based on CRC32 hash
def test_set_check(identifier, test_ratio):
# Calculate hash value and assign based on the test ratio
return crc32(identifier.encode()) & 0xffffffff < test_ratio * 2**32
# Function to split the dataset using the 'persistent_id' column
def split_train_test_by_id(data, test_ratio, id_column):
ids = data[id_column].astype(str) # Convert IDs to string if necessary
in_test_set = ids.apply(lambda id_: test_set_check(id_, test_ratio)) # Apply hash check
return data.loc[~in_test_set], data.loc[in_test_set] # Return train and test sets
# Split the male-only data into 80% train and 20% test sets using 'persistent_id'
train_set, test_set = split_train_test_by_id(male_straight_data_with_persistent_id, 0.2, "persistent_id")
# Display the sizes of the train and test sets
print(f"Training set size: {train_set.shape[0]}")
print(f"Test set size: {test_set.shape[0]}")
# Save the train and test sets as CSV files
train_set.to_csv(os.path.join(DATING_PATH, "male_straight_data_with_persistent_id_train_set.csv"), index=False)
test_set.to_csv(os.path.join(DATING_PATH, "male_straight_data_with_persistent_id_test_set.csv"), index=False)
print("Train and test sets have been successfully saved.")Through this deep dive into the OkCupid dataset, we've laid the groundwork for predicting mating success for straight males by carefully refining our feature set and using persistent IDs for stable train-test splits. We've explored essential concepts, from filtering relevant data to calculating CRC32 hashes, and applied these methods to ensure consistent results across multiple runs.
Our next steps will focus on applying machine learning models to uncover patterns between body type, ethnicity, height, and personal habits, aiming to predict which factors play the most significant role in dating success.
To get the full picture of this project, I recommend having a read of the following blog posts:
Let's use an example
UUID:
d8ea2885-a97e-4400-bc52-cf093c343e10
This UUID is in hexadecimal format (base-16). Each two-hex character represents one byte (or eight bits). Let's convert these values.
Hexadecimal values use base-16, meaning each digit can be from 0 to F (where A = 10, B = 11, ..., F = 15).
d in hex = 13 (decimal)
8 in hex = 8 (decimal)
Each position in hexadecimal corresponds to a power of 16.
\[ d8_{16} = (13 \times 16^1) + (8 \times 16^0) \]
\[ d8_{16} = (13 \times 16) + (8 \times 1) = 208 + 8 = 216 \]
Thus, d8 in hex equals 216 in decimal.
Let's now convert all parts of the UUID into decimal values and byte (binary) values.
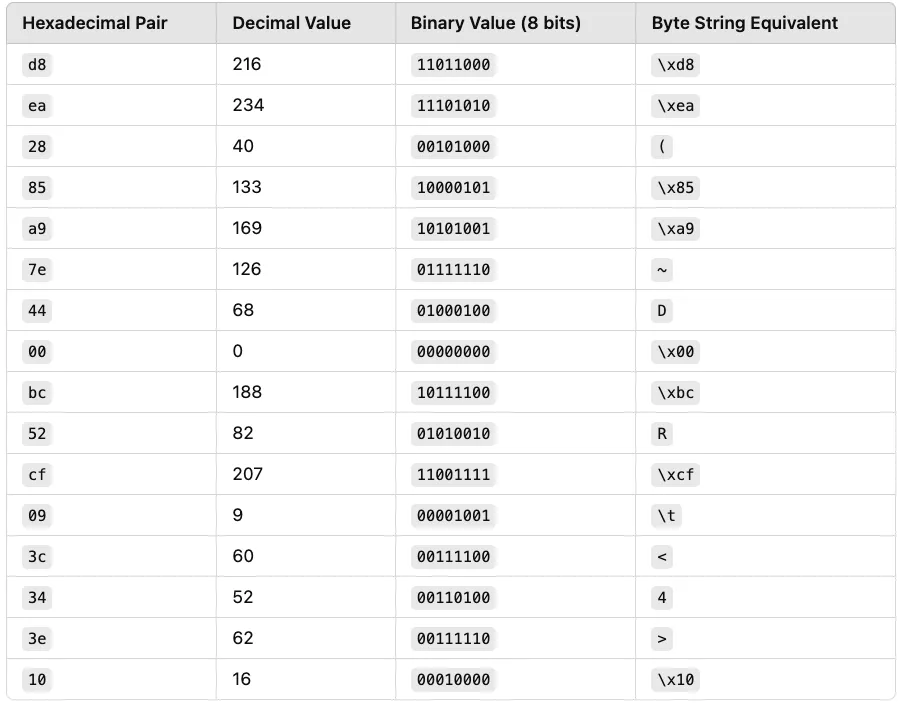
Once converted, the UUID becomes the following 16-byte string:
b'\xd8\xea(\x85\xa9~D\x00\xbcR\xcf\t
This byte string is the format needed for the CRC32 hashing function.
Now, let's compute the CRC32 hash of the byte string step-by-step.
CRC32 stands for Cyclic Redundancy Check. A polynomial division algorithm produces a 32-bit integer hash from input data (in our case, the UUID byte string).
The output is a 32-bit hash, ranging from 0 to 2^32 - 1 (4,294,967,295).
from zlib import crc32
# Compute CRC32 hash of the byte string
byte_string = b'\xd8\xea(\x85\xa9~D\x00\xbcR\xcf\t\x10'
hash_value = crc32(byte_string)
print(hash_value) # Example output: 2441647152 When CRC32 computes a hash, it may return a signed 32-bit integer (which can be negative). However, we want to ensure the hash is treated as an unsigned 32-bit integer.
unsigned_hash = hash_value & 0xffffffff
0xffffffff is the largest 32-bit unsigned integer:
0xffffffff = 4,294,967,295 (decimal)
The & (bitwise AND) operation keeps only the lower 32 bits of the hash, ensuring it's treated as unsigned.
2441647152 & 0xffffffff = 2441647152
Since the hash is already positive, this operation doesn't change its value.
Let's assume the test ratio is 0.2 (20%).
\[ \text{Threshold} = 0.2 \times 2^{32} = 0.2 \times 4,294,967,296 = 858,993,459.2 \]
We compare the hash value with the threshold:
2441647152 < 858993459 # False
Since 2441647152 is greater than the threshold, this entry does not belong to the test set. It will be assigned to the training set.